Abstract
Background
Diffuse large B-cell lymphoma (DLBCL) is a genetically and clinically heterogeneous disease. The cell-of-origin (COO) classification subdivides DLBCL into the transcriptionally defined activated B-cell (ABC) and germinal center B-cell (GCB) subtypes. While RNA based methods are considered the gold standard to determine COO, they are rarely used in clinical routine due to logistical and methodological challenges. Alternatives include the immunohistochemistry-based Hans algorithm and classifiers to infer the COO subtype from DNA sequencing [Esfahani et al., Blood 2019]. Despite the undisputed value of these methods, the concordance with the gold standard RNA and their prognostic implication are limited. We have recently shown that expression of individual genes can be inferred from cfDNA fragmentation patterns using a method called EPIC-Seq (EPigenetic expression Inference from Cell-free DNA Sequencing) [Esfahani et al., Cancer Res. 2020]. We therefore reasoned that EPIC-Seq may improve COO classification compared to other non-RNA methods.
Methods
A gene expression model, using cfDNA fragmentation patterns, was trained using leukocyte RNA-sequencing and deep whole genome profiling of the plasma cell-free DNA of an individual with no evidence of circulating disease. The trained model takes two features into account to infer gene expression: 1. promoter fragmentation entropy (PFE), and 2. normalized coverage at the nucleosome-depleted region of a given transcription start site (TSS). We then used a capture panel targeting TSS specifically designed for EPIC-Seq. We selected genes based on their power to discriminate COO subtypes in tumor RNA sequencing [Schmitz et al., NEJM 2018]. We first developed a classifier to distinguish DLBCL cases from healthy plasma using the inferred gene expression. Moreover, we defined GCB and ABC signature scores as the average inferred expression of a set of 'GCB genes' (n=34) and 'ABC genes' (n=34), respectively. Finally, we defined the COO score as the difference between the GCB and ABC scores. To validate our assay and method, we profiled 71 plasma samples from 68 healthy individuals and 90 pretreatment plasma samples from patients with large B-cell lymphomas using EPIC-Seq.
Results
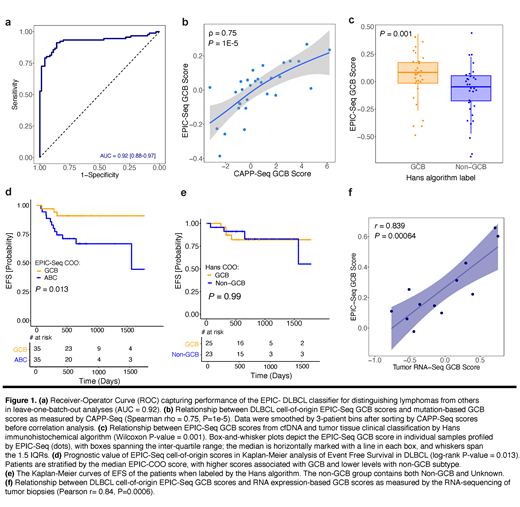
We first evaluated the performance of the EPIC-Seq classifier in distinguishing DLBCL cases from controls and achieved an AUC of 0.92 in a cross-validation setting (Fig 1a). We then compared the result of EPIC-Seq COO classifier with the genotype-based method previously developed in our group. We observed epigenetic scores to be significantly correlated with previously described mutation-based GCB scores (r=0.75, P=1E-5, Fig. 1b). When comparing to the Hans classification algorithm, we observed significantly higher GCB scores in cases classified as GCB by Hans as compared with non-GCB cases (Wilcox P=0.001, Fig. 1c). Comparing the prognostic power of epigenetic and mutation-based COO labels in previously untreated patients using univariate Cox regressions, the EPIC-Seq classifier better stratified event-free survival (EFS) with higher GCB scores being associated with favorable outcomes (n=70, EPIC-Seq: HR=0.13, P=0.033 vs CAPP-Seq: HR=0.95, P=0.62). Importantly, when binarizing patients into GCB and non-GCB cases by the median, patients with tumors classified as GCB had significantly longer EFS than non-GCB counterparts (log-rank P=0.013, Fig. 1d). The Hans algorithm, in contrast, failed to stratify patients for EFS, among patients analyzed by both immunohistochemistry and DNA genotyping (Fig. 1e). Finally, we profiled n=12 additional DLBCL cases by both RNA-sequencing and EPIC-Seq. Strikingly, we found EPIC-COO scores to be significantly correlated with RNA based GCB scores (r=0.84, P=6E-4, Fig. 1f) underscoring the concordance of EPIC-Seq based COO classification with a gold standard scoring system.
Conclusions
We here apply EPIC-Seq, a method to infer expression of individual genes from cfDNA fragmentation patterns, to classify DLBCL patients into COO subtypes. COO classification by EPIC-Seq outperformed both Hans and mutation-based methods with regards to outcome stratification and correlated well with RNA-based methods. Overall, these results suggest that EPIC-Seq has utility for noninvasive classification of DLBCL cell-of-origin subtypes and may help to overcome logistical and methodological challenges of RNA-based methods.
Shahrokh Esfahani: Foresight Diagnostics: Current holder of stock options in a privately-held company. Kurtz: Genentech: Consultancy; Foresight Diagnostics: Consultancy, Current holder of stock options in a privately-held company; Roche: Consultancy. Diehn: Foresight Diagnostics: Current holder of individual stocks in a privately-held company, Current holder of stock options in a privately-held company; CiberMed: Current holder of stock options in a privately-held company, Patents & Royalties; Illumina: Research Funding; Varian Medical Systems: Research Funding; BioNTech: Consultancy; RefleXion: Consultancy; AstraZeneca: Consultancy; Roche: Consultancy. Alizadeh: Gilead: Consultancy; Bristol Myers Squibb: Research Funding; Janssen Oncology: Honoraria; Celgene: Consultancy, Research Funding; Forty Seven: Current holder of individual stocks in a privately-held company, Current holder of stock options in a privately-held company; CAPP Medical: Current holder of individual stocks in a privately-held company, Current holder of stock options in a privately-held company; Roche: Consultancy, Honoraria; Foresight Diagnostics: Consultancy, Current holder of individual stocks in a privately-held company, Current holder of stock options in a privately-held company; Cibermed: Consultancy, Current holder of individual stocks in a privately-held company, Current holder of stock options in a privately-held company.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal